March 14, 2023
RSV vaccines: the latest success story
After decades of research, structure-based design efforts are yielding effective vaccines against respiratory syncytial virus. Jason McLellan explains the breakthroughs that paved the way.
Kristen Kresge Abboud
Respiratory syncytial virus (RSV) seemed an easy foe. Not long after the virus was isolated from a chimpanzee with a respiratory infection and determined to be of human origin, vaccine development efforts were underway (see timeline on RSV vaccine development below).
Approximately 64 million people worldwide suffer from respiratory tract infections caused by RSV every year, but the virus is most serious in the very young and the elderly. Nearly 3 million children under the age of five and more than 300,000 older adults are hospitalized annually due to RSV infection, which in some cases is fatal. RSV poses an even greater risk of death to children under one year of age than influenza.
But whatever optimism there was that a vaccine would be readily developed faded quickly in the mid-1960s when a whole-killed RSV vaccine candidate led to enhanced disease among infants who were subsequently exposed to naturally circulating virus, killing two children and sending others to the hospital. Following that, the focus shifted to understanding the immunological determinants of what had gone wrong, what researchers came to refer to as RSV-vaccine-associated enhanced respiratory disease.
Then, in 2013, a critical discovery put researchers on a new track to developing an RSV vaccine, this time backed by structure-based vaccine design. This discovery was made by Jason McLellan, a structural biologist who at the time was completing his postdoctoral studies at the Vaccine Research Center (VRC) at the National Institute of Allergy and Infectious Diseases (NIAID) at the U.S. National Institutes of Health (NIH).

McLellan came to the VRC and began working on HIV but soon shifted his focus to RSV when he teamed up with veteran infectious disease researcher Barney Graham, who had already been working on RSV for decades by that time.
Working backwards from an antibody to RSV, McLellan and colleagues identified sites on the surface protein of the virus, known as F, that are particularly vulnerable to attack by neutralizing antibodies, making it an ideal vaccine immunogen.
The problem was that the F protein was very unstable, flipping easily from one conformation to another, and it was only in one specific conformation, known as prefusion, that these highly desirable parts of the protein known as epitopes were exposed. Once the F protein is in the postfusion state, these epitopes are hidden.
To engineer the F protein as a vaccine immunogen, researchers therefore needed to lock it in the prefusion conformation. In 2013, McLellan and colleagues reported that they did exactly that. It ended up being one of the scientific breakthroughs of the year in Science magazine, where editors had this to say: “Researchers have long hoped that structural biology, the study of the molecules of life, would help them design better vaccines. This year, it began to deliver.”
This discovery formed the basis for many of the RSV vaccine candidates that are now in development, two of which may soon be authorized by the U.S. Food and Drug Administration — one from Pfizer, the other from GSK. It also led to the development of the stabilized Spike protein of SARS-CoV-2 that was used in the first COVID-19 vaccines.
Where it will lead to next is anybody’s guess.
You might say McLellan, now a professor at the University of Texas at Austin, has a knack for solving structures, and it turns out that these structures are giving way to incredibly important vaccines. I spoke with McLellan about the arc of RSV vaccine development and how structural biology and machine learning together are heralding a new era of vaccines.
An edited version of our conversation appears below.
What was the key to unlocking the prefusion conformation of the RSV F protein and why was it so difficult to do?
It was difficult just because the RSV F protein is so unstable. The prefusion conformation is what is referred to as meta-stable, which means it is not its lowest energy conformation, and so there is a kinetic barrier keeping it from staying in this state. For some viral fusion proteins this barrier can be quite high, and RSV may be one of the most unstable viral fusion proteins in its class. Even just purifying the virus triggers it to go into the postfusion conformation, but we don’t actually know what the specific trigger is. For HIV, for example, HIV Envelope binding to its receptor on cells induces a conformational change, but for RSV we don’t know what the receptor is or even if there is one for the RSV F protein.
The F protein is just very unstable, and to stabilize it in its prefusion state we first needed to come up with some method of obtaining a reasonably high resolution structure that we could use to engineer in amino acid substitutions to help stabilize it in that conformation. And to do that, the trick was to identify monoclonal antibodies that could bind exclusively to the prefusion conformation and prevent the protein refolding from occurring.
The fact that the receptor for the F protein, if there is one, isn’t known makes it sound as though there are some gaps in our understanding of the virology of RSV. Is that true?
There are a lot of gaps in the virology. I think it’s kind of amazing that we’ll have a vaccine before we really figure out the molecular details of entry for the virus.
How did you identify the antibodies to RSV that allowed you to do the structural work that was required?
It started with a really nice paper in 2011 from Jose Melero and colleagues. They took serum from people that had high titers of RSV-neutralizing antibodies and passed it over a column mobilized with the postfusion form of F. Antibodies specific to the postfusion form would stick to the column, but what they found was that most of the neutralizing antibodies passed through the column without sticking, suggesting they bound to some other conformation. That paper was key.
Based on that, we set out to obtain one or more prefusion-specific binding antibodies, and we eventually found one mouse antibody and one human antibody that bound only the prefusion conformation, not the postfusion.
The next step was to trap one of these antibodies in complex with the prefusion F. To do that, I expressed the unstable F protein in the presence of the human antibody called D25. The idea was that the antibody would bind, lock it in place, and then we could purify the complex of an antibody-bound prefusion protein, and this actually worked really well.
A derivative of this antibody, Nirsevimab, will likely be approved this year as a prophylactic antibody for infants.
Then you needed to identify the epitopes on the prefusion F that were susceptible to antibodies, right?
Yes. We knew that all the antibodies to the prefusion F protein bound to the apex. This was a novel site that nobody had seen before, in part because nobody had identified the prefusion F structure, and this is the region that completely refolds into a different conformation in the postfusion form so immunizing with postfusion F is never going to elicit these antibodies. The goal then was to use structure-based design to engineer amino acid substitutions into the F protein that help lock it in the prefusion state and prevent the conformational change from occurring.
This worked too, and when you immunized animals, you induced neutralizing antibodies that could protect, which makes me think about HIV. Even after stabilizing the HIV Envelope protein it still doesn’t induce the types of neutralizing antibodies that are protective. Luckily that wasn’t the case with RSV.
We’ve learned a lot of lessons from RSV, but they don’t help overcome the issues plaguing HIV vaccine development. When I joined Peter Kwong’s lab at the VRC, the whole lab was focused on HIV vaccine design and structure-based vaccine design, and I thought we were coming up with cool ideas for protein stabilization and epitope scaffolding. But nothing worked. And when that happens it’s hard to know if the ideas are flawed, or if HIV is just such a difficult virus. So I decided to try some of these approaches with a more tractable virus and Barney suggested that RSV was a perfect test case. And it was a terrific test case to show that structure-based vaccine design can work.
Trying to create an HIV vaccine is incredibly difficult but in the process of trying to do so you are going to spin off cool technologies. Many other fields have benefitted from the advances in structure-based design, B-cell sorting, and antibody isolation that were pioneered for HIV.
Which is what happened with COVID also.
That’s right. We published two papers on the proof of concept for structure-based vaccine design with RSV in 2013 and this work was one of Science magazine’s top 10 breakthroughs of the year. We thought RSV was going to be the first vaccine developed based on this work, but, of course, it took a decade to actually develop the vaccine candidates and COVID came along in the meantime.
Fortunately, we were prepared for that because after RSV we started working on the Middle East respiratory syndrome (MERS) coronavirus. We couldn’t initially obtain the structure of the MERS Spike protein, but we did obtain a structure of the Spike protein from another human coronavirus, HKU1. Using this structure, we engineered stabilizing substitutions that worked for MERS Spike, as well as related betacoronavirus Spikes. These substitutions also increased Spike expression, and this meant we were well prepared to do the same thing when SARS-CoV-2 emerged, which is partly why vaccines were developed so rapidly.
So where do you go next?
Basically everywhere — the sky’s the limit. And it’s not just viruses. People are starting to do the same thing now for bacterial pathogens and parasites, such as malaria. It’s not going to solve all the problems, but I think it’s clearly become a very important tool for vaccine developers, along with all the new advances in machine learning. It’s a super exciting time.
How could machine learning or AlphaFold help you with your work on designing vaccine immunogens?
I think there’s a lot of enthusiasm about sort of running AlphaFold backwards, if you will. For AlphaFold the input is a sequence, and the output is a protein structure. We’re interested in the input being a protein structure and the output being a sequence that would help us identify amino acid substitutions that would stabilize the structure.
Right now, we do almost all of it by eye or by using some programs, but we might have to design 100 stabilizing substitutions and test them all individually to get 20 or 25 that work. We’d love it if machine learning could cut that down to testing 50 and getting 25 that work.
And what’s even more exciting would be if a machine-learning program can come up with substitutions we haven’t even thought of through things it has learned. There’s a lot of excitement with everything that’s going on with that. I think it’s a peak time in the next 10 years where we will get a burst of new vaccines based on these new technologies.
Persistence pays
A timeline of the key scientific progress in RSV vaccine development
 Creative artwork featuring 3D renderings of RSV — a common contagious virus that infects the human respiratory tract. Credit: NIAID/NIH
Creative artwork featuring 3D renderings of RSV — a common contagious virus that infects the human respiratory tract. Credit: NIAID/NIH
1956: Respiratory syncytial virus (RSV) is isolated from a lab animal with an upper respiratory tract disease. The paramyxovirus is initially referred to as “chimpanzee coryza agent.”
1957: RSV is quickly determined to be of human origin and is found to be the cause of a common lung infection in young children and infants.
1965-1967: Following on the success of the whole, inactivated polio vaccine developed by Jonas Salk, researchers develop a whole, inactivated RSV vaccine candidate and test it in four large studies. The vaccine is not efficacious, and, in fact, causes enhanced disease in infants subsequently infected with naturally circulating RSV, temporarily derailing RSV vaccine research efforts.
1982: The viral genome of RSV is sequenced.
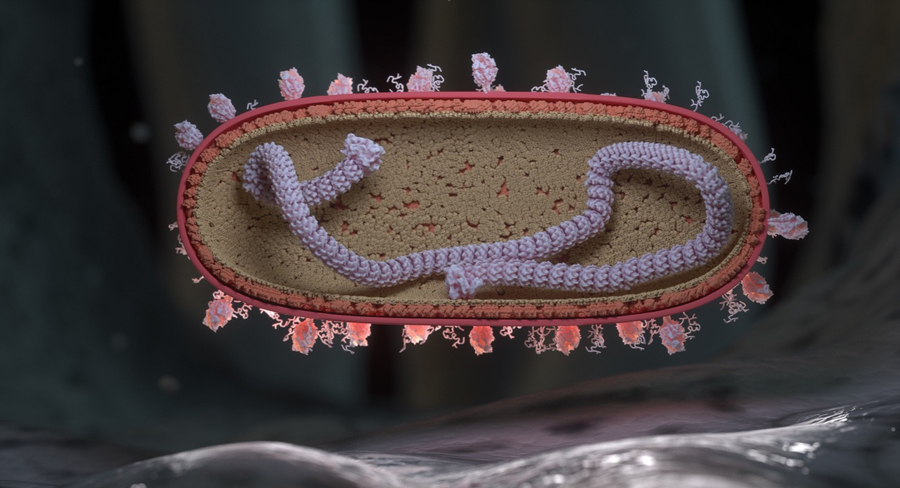 RSV virion in cross-section landing on the surface of a human respiratory endothelial cell. Single stranded RNA (colorized purple) is shown wrapped around a nuclear protein core capped by large L proteins. M2-1 and M proteins line the inside of the virion and F and G proteins populate the outer surface of the virion. F proteins are represented as drumstick like structures and G proteins are represented as tangled tubular structures. Credit: NIAID/NIH
RSV virion in cross-section landing on the surface of a human respiratory endothelial cell. Single stranded RNA (colorized purple) is shown wrapped around a nuclear protein core capped by large L proteins. M2-1 and M proteins line the inside of the virion and F and G proteins populate the outer surface of the virion. F proteins are represented as drumstick like structures and G proteins are represented as tangled tubular structures. Credit: NIAID/NIH
1983: Researchers isolate mouse monoclonal antibodies to RSV.
1984: The surface glycoprotein and fusion glycoprotein (F) of the virus are isolated.
1986: Recombinant RSV proteins are derived from poxvirus vectors.
1989: Recombinant RSV F protein is derived from insect cells.
1990s/early 2000s: Small animal models are used to elucidate the immunological determinants of RSV-vaccine-associated enhanced respiratory disease. At the same time, researchers expand their understanding of the role of T cells in RSV pathogenesis.
Following this, vaccine research efforts are “cautiously reinitiated.” Five Phase III trials of vaccine candidates based on the prefusion conformation of the RSV F protein are conducted but none are efficacious enough.
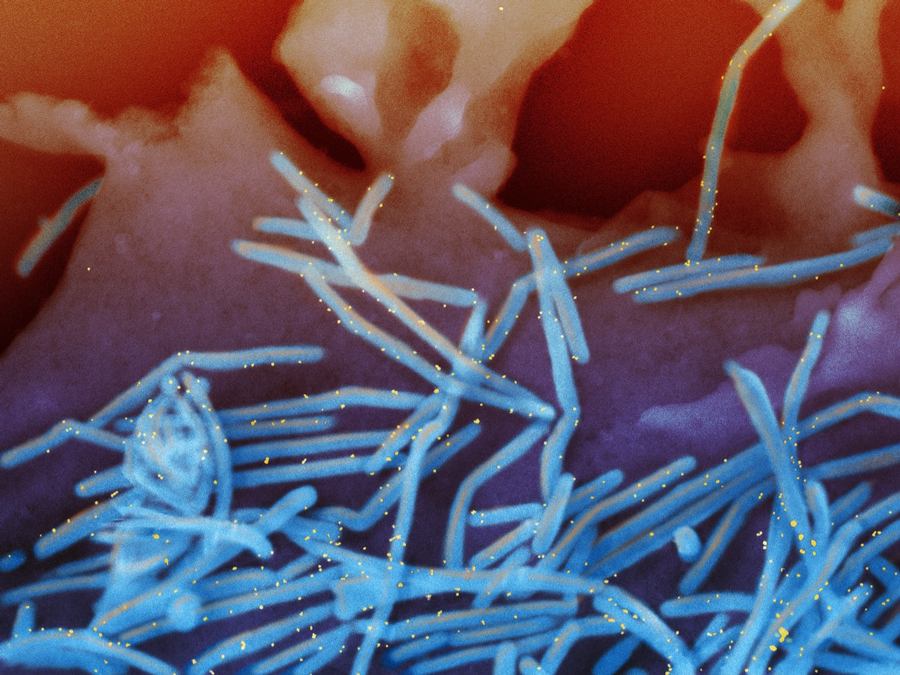 Scanning electron micrograph of RSV virions (colorized blue) and labeled with anti-RSV F protein/gold antibodies (colorized yellow) shedding from the surface of human lung epithelial A549 cells. Credit: NIAID/NIH
Scanning electron micrograph of RSV virions (colorized blue) and labeled with anti-RSV F protein/gold antibodies (colorized yellow) shedding from the surface of human lung epithelial A549 cells. Credit: NIAID/NIH
1997: Researchers show that passive administration of a human monoclonal antibody known as palivizumab can protect against severe disease from RSV.
2006: The first atomic-level structure is published of a paramyxovirus F protein in its prefusion conformation.
2012: Researchers show that a major target of antibodies induced by natural RSV infection is on the prefusion conformation of the virus’s F protein, making this less stable form of the viral protein an obvious target for vaccine design efforts.
2013: McLellan and colleagues at the VRC at NIAID at the NIH create a stabilized version of the RSV F protein in its prefusion conformation, kicking off vaccine development efforts based on the prefusion F protein.
 Engineering a stable prefusion RSV protein as a vaccine antigen. The left drawing shows the trimeric RSV F protein in its unstable, prefusion state. The bulb-like protein is anchored to the viral envelope at the C-terminus. At the top of the F protein in its prefusion conformation are the epitopes (in red) that are targeted by antibodies with high neutralizing activity. In the postfusion conformation of the F protein, these epitopes are no longer exposed and are therefore unable to be targeted by vaccine-induced antibodies. Credit: Based on the image in N. Engl. J. Med. 2023; 388:579-58.
Engineering a stable prefusion RSV protein as a vaccine antigen. The left drawing shows the trimeric RSV F protein in its unstable, prefusion state. The bulb-like protein is anchored to the viral envelope at the C-terminus. At the top of the F protein in its prefusion conformation are the epitopes (in red) that are targeted by antibodies with high neutralizing activity. In the postfusion conformation of the F protein, these epitopes are no longer exposed and are therefore unable to be targeted by vaccine-induced antibodies. Credit: Based on the image in N. Engl. J. Med. 2023; 388:579-58.
2019: Structure-based vaccine design work used to stabilize the RSV F protein in its prefusion conformation are applied to coronaviruses in response to outbreaks of Middle East respiratory syndrome (MERS).
2020: When SARS-CoV-2 emerges, researchers use similar techniques identified through work on RSV to stabilize the Spike protein of the virus, leading to the development of highly efficacious COVID-19 vaccines in record time.
2023: Advisers to the U.S. Food and Drug Administration recommend that the agency approve two RSV vaccine candidates for older adults, one developed by Pfizer and the other by GSK.
Sources:
- The Journey to RSV Vaccines, Barney S. Graham, New England Journal of Medicine.
- Structure-Based Design of a Fusion Glycoprotein Vaccine for Respiratory Syncytial Virus, McLellan et. al., Science; 2013 November 1; 342(6158): 592–598.
- Respiratory syncytial virus vaccine development, Julia L. Hurwitz, Expert Review Vaccines; 2011 October; 10(10): 1415-1433.